 www.revisionesencancer.com
www.revisionesencancer.com
1 )
“Tratamiento médico del cáncer en el año 2019”
Organiza:
6 , 7 y 8 d e f e b r e r o 2 0 1 9
NH Collection Eurobuilding
M A D R I D
www.revisionesencancer.comC O O R D I N A D O R E S C I E N T Í F I C O S :
Eduardo Díaz-Rubio
Enrique Aranda Aguilar
Enrique Grande Pulido
Ana Lluch Hernández
Pedro Pérez Segura
Mariano Provencio Pulla
¿Son todos los test genómicos iguales?
CLINICAL TRIAL
Prognostic ability of EndoPredict compared to research-based
versions of the PAM50 risk of recurrence (ROR) scores in node-
positive, estrogen receptor-positive, and HER2-negative breast
cancer. A GEICAM/9906 sub-study
Miguel Martin
1
•
Jan C. Brase
2
•
Amparo Ruiz
6
•
Aleix Prat
7
•
Ralf Kronenwett
2
•
Lourdes Calvo
8
•
Christoph Petry
2
•
Philip S. Bernard
9
•
Manuel Ruiz-Borrego
10
•
Karsten E. Weber
2
•
Ce´sar A. Rodriguez
11
•
Isabel M. Alvarez
12
•
Miguel A. Segui
13
•
Charles M. Perou
3,4,5
•
Maribel Casas
14
•
Eva Carrasco
14
•
Rosalı´a Caballero
14
•
Alvaro Rodriguez-Lescure
15
Received: 30 December 2015/Accepted: 16 February 2016/Published online: 24 February 2016
!
The Author(s) 2016. This article is published with open access at Springerlink.com
Abstract
There are several prognostic multigene-based
tests for managing breast cancer (BC), but limited data
comparing them in the same cohort. We compared the
prognostic performance of the EndoPredict (EP) test (s-
tandardized for pathology laboratory) with the research-
based PAM50 non-standardized qRT-PCR assay in node-
positive estrogen receptor-positive (ER
?
) and HER2-
negative (HER2
-
) BC patients receiving adjuvant
chemotherapy followed by endocrine therapy (ET) in the
GEICAM/9906 trial. EP and PAM50 risk of recurrence
(ROR) scores [based on subtype (ROR-S) and on subtype
and proliferation (ROR-P)] were compared in 536 ER
?
/
HER2
-
patients. Scores combined with clinical informa-
tion were evaluated: ROR-T (ROR-S, tumor size), ROR-
PT (ROR-P, tumor size), and EPclin (EP, tumor size, nodal
status). Patients were assigned to risk-categories according
to prespecified cutoffs. Distant metastasis-free survival
Electronic supplementary material
The online version of this
article (doi:
10.1007/s10549-016-3725-z
) contains supplementary
material, which is available to authorized users.
&
Miguel Martin
mmartin@geicam.org1
Department of Medical Oncology, Instituto de Investigacio´n
Sanitaria Gregorio Maran˜on, Universidad Complutente de
Madrid, Calle Maiquez 7, Madrid, Spain
2
Sividon Diagnostics GmbH, Cologne, Germany
3
Lineberger Comprehensive Cancer Center, University of
North Carolina, Chapel Hill, NC, USA
4
Department of Genetics, University of North Carolina,
Chapel Hill, NC, USA
5
Department of Pathology & Laboratory Medicine, University
of North Carolina, Chapel Hill, NC, USA
6
Department of Medical Oncology, Valencian Institute
of Oncology (IVO), Valencia, Spain
7
Translational Genomics Group, Vall d’Hebron Institute
of Oncology (VHIO), Barcelona, Spain
8
Department of Medical Oncology, A Corun˜a University
Hospital Complex, A Corun˜a, Spain
9
Solid Tumor Molecular Diagnostics Laboratory, ARUP
Laboratories, Utah, USA
10
Department of Medical Oncology, Virgen del Rocio
University Hospital, Seville, Spain
11
Department of Medical Oncology, Salamanca University
Hospital-IBSAL, Salamanca, Spain
12
Department of Medical Oncology, Donostia University
Hospital, Donostia, Spain
13
Department of Medical Oncology, Parc Tauli University
Hospital, Sabadell, Spain
14
Spanish Breast Cancer Research Group (GEICAM), Madrid,
Spain
15
Department of Medical Oncology, Elche University General
Hospital, Elche, Spain
123
Breast Cancer Res Treat (2016) 156:81–89
DOI 10.1007/s10549-016-3725-z
84
Breast Cancer Res Treat (2016) 156:81–89
predictors would not improve prognostic performance.
These findings are concordant with our previous combined
analysis of hundreds of signatures and clinical-pathological
data for prognostic prediction in ER-positive breast cancer
where we observed that not much more prognostic power
was obtained by including hundreds of signatures into a
single model beyond the power contained within a well-
developed individual signature when combined with clin-
ical variables [
28
].
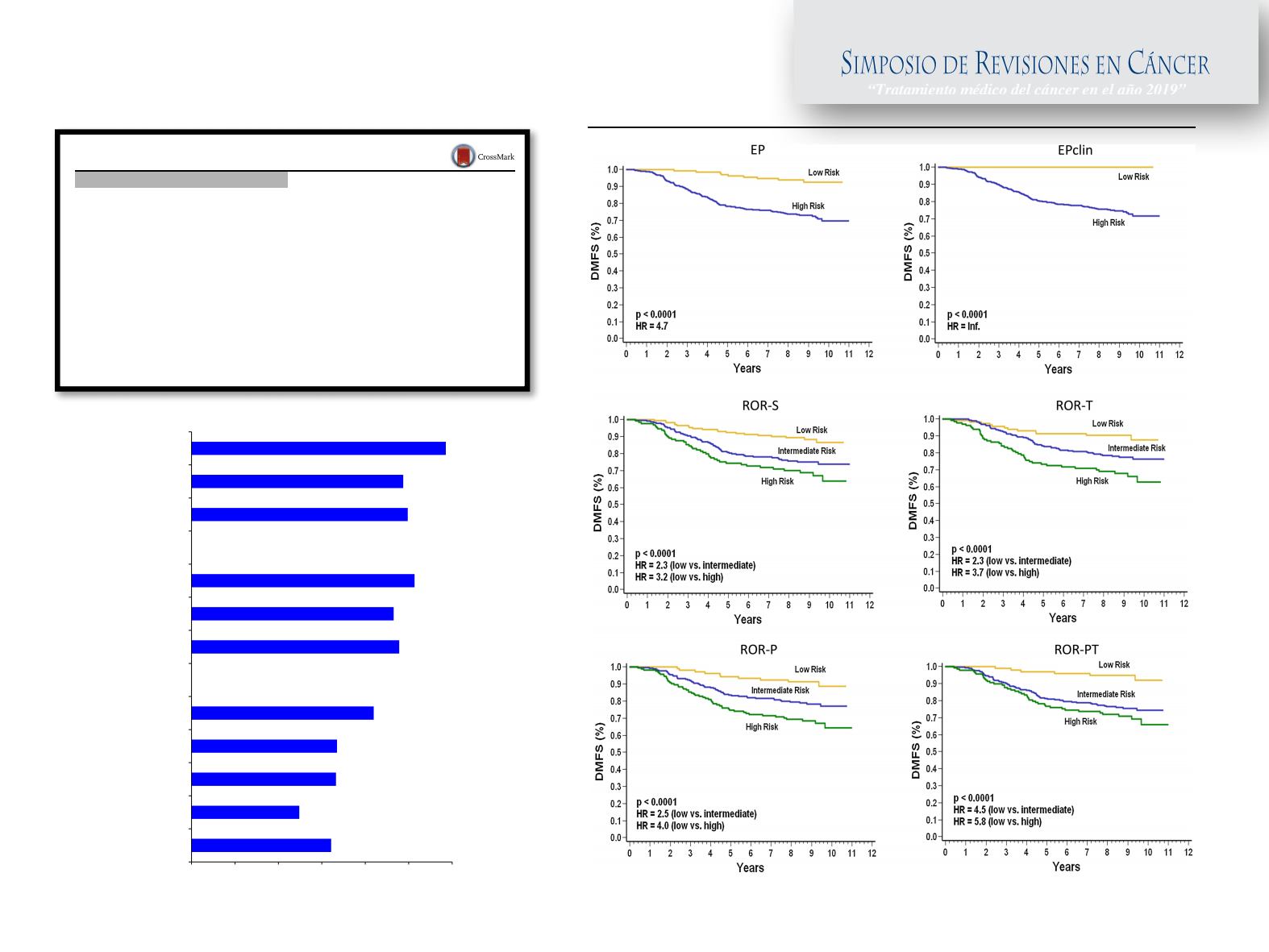
The PAM50-based ROR-T and ROR-PT scores include
tumor size, whereas the EPclin score considers nodal status
and tumor size, as part of the risk prediction algorithm.
Similar to the research-based version, a ROR-PT score
weighted for tumor size and proliferation was used to
validate the standardized version of PAM50 assay in the
ATAC and ABCSG8 trials. In our analysis, all hybrid
scores contributed to identifying low-risk groups for distant
metastasis, although number of patients and events differed
across score categories. The EPclin low-risk group was
smaller than the ROR-T and ROR-PT ones and showed no
distant-metastatic events. EPclin had been established in a
node-positive/node-negative cohort and the predefined cut-
off level consequently classified more patients as high-risk
in the node-positive GEICAM/9906 trial. In contrast, the
research-based versions of ROR-T and ROR-PT scores
were derived in a systemically untreated node-negative BC
cohort, and thresholds were based on subtype distribution
and not actual survival outcomes; therefore, the number of
0.4 0.45 0.5 0.55 0.6 0.65 0.7
Age
Arm
Grade
Tumor Size
Nodal Status
ROR-S
ROR-P
EP
ROR-T
ROR-PT
EPclin
C-index
Fig. 3
Distribution of clinical and molecular parameters c-indices.
EP
EndoPredict score,
EPclin
EP based on tumor size and nodal
86
Breast Cancer Res Treat (2016) 156:81–89








